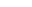Genomics and Bioinformatics of Horticulture Plants
Horticulture plants are vitally important to human society providing food, nutrients, medicine, aroma and ornament to our everyday life. Mapping the genetic codes of horticulture plants and understanding their functional and regulatory mechanisms are key to developing technologies and products for conservation and utilization of these high-value crops. Our group uses the state-of-the-art genome sequencing and analytical techniques to assemble, annotate and analyze the genomes of major horticulture plants including vegetables, flowers, fruits as well as medicinal herbs. Genomic studies of these plants will provide an essential foundation and resources to improve crops yield, quality and resistance to pests using molecular breeding and modern biotechnologies.
Molecular basis of crop resistance to Fusarium vascular wilt
Fusarium wilt is a common soil-borne disease of over 100 different horticulture plants including tomato, cabbage, watermelon, banana, pepper etc. caused by filamentous fungus Fusarium oxysporum. Resistant cultivars against Fusarium wilt are quite limited due to our poor understanding of plant immunity and fungal pathogenic mechanisms. Chemical control of the diseases is often cost-ineffective and unfriendly to environment. Our group uses Arabidopsis, tomato and watermelon as model species to dissect the molecular mechanisms of host and F. oxysporum interactions, with the goal of identifying key genetic and epigenetic components critical for pathogenesis and immunity.
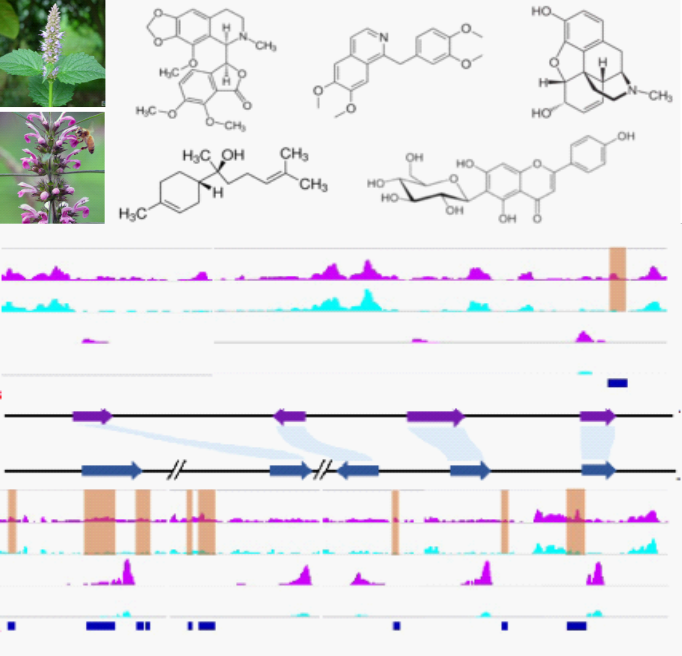
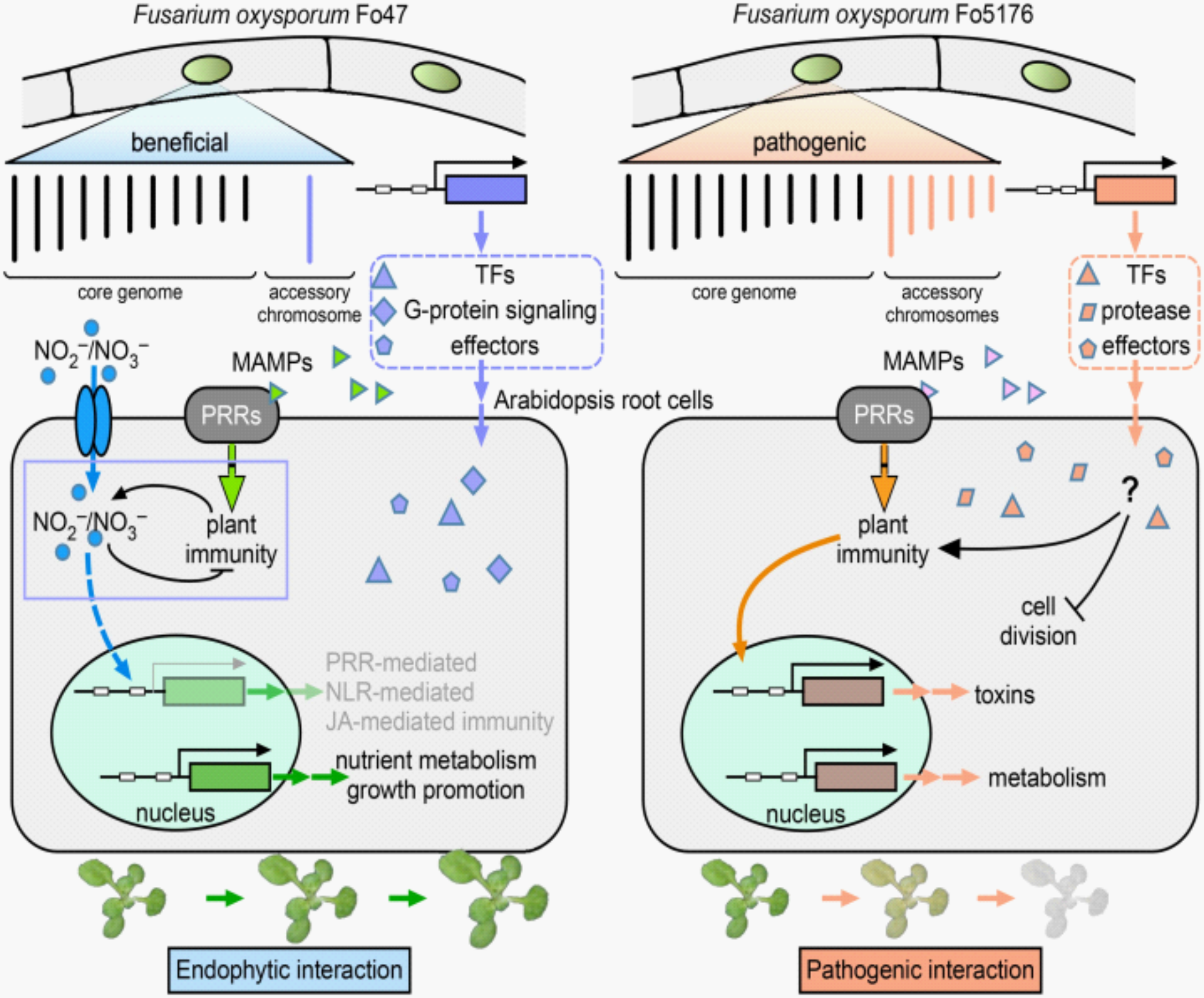
Synthetic Biology of Plant and Fungal Natural Products
Natural products are secondary metabolites produced by plants and microorganisms with diverse chemical structures and pharmaceutical properties used to treat various human diseases. However, most of these natural substances are hard to manufacture chemically given their complex structures and biosynthetic pathways. Using a combination of genomics, transcriptomics and metabolomics data mining, we sought to elucidate the full biosynthetic pathways of important plant and fungal natural products, followed by metabolic engineering of these pathways in heterologous hosts (yeast and tobacco). Alternatively, we study the regulatory mechanisms underlying the biosynthesis in plants and fungi and use the knowledge to guide molecular breeding of plants with high yield of these chemicals using transgenic and genome editing methods.

Functional characterization of secondary metabolic gene clusters
Metabolic gene clusters are operon-like gene structures present in eukaryotic cells including fungi and plants, largely responsible for production of secondary metabolites. Our group is particularly interested in plant and fungal gene clusters that play active roles in mediating interactions of plants and pathogens. Many of these putative gene clusters have no known biological functions and we aim to study their structure, function and regulatory mechanisms using a combination of genomic, molecular genetic and biochemical approaches.